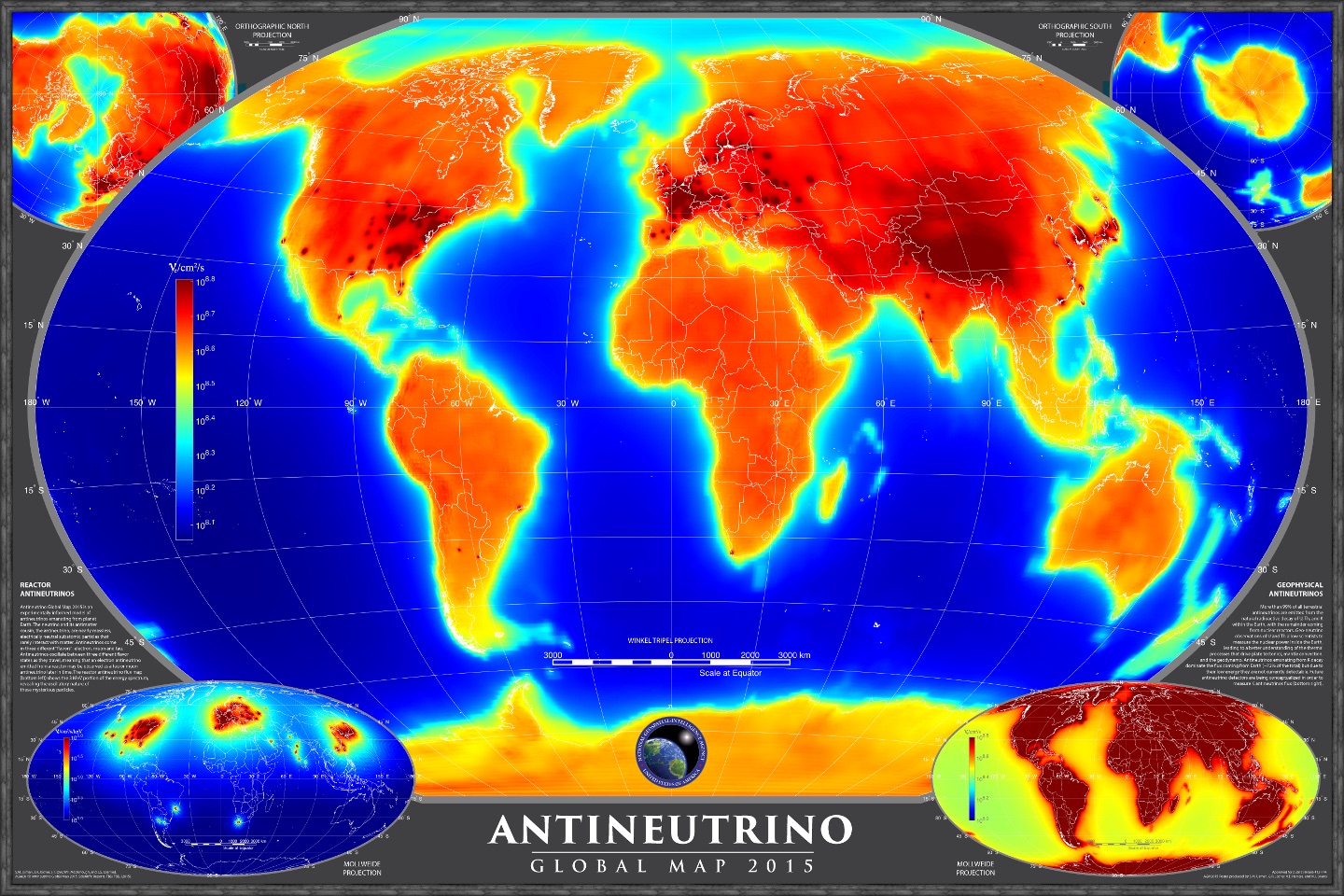
Welcome to the AntiNeutrino Global Map (AGM) 2015 production code repository. This project represents an innovative endeavor by Ultralytics to visualize and analyze antineutrino emissions across the globe, contributing valuable insights to fields like geoscience and particle physics. The map produced by this code provides a unique view of antineutrino emissions, capturing data from both natural and artificial sources, showcasing the power of data visualization in scientific research.
This repository hosts the production code for AGM2015, an impactful scientific study detailed in the paper "AGM2015: Antineutrino Global Map 2015," published in Scientific Reports. This research enhances our understanding of antineutrino emissions and their distribution around the Earth.
- Paper: S.M. Usman, G.R. Jocher, S.T. Dye, W.F. McDonough, and J.G. Learned - Read the paper on Nature Scientific Reports
For additional context on the study's significance, you can refer to the National Geospatial-Intelligence Agency (NGA) Press Release:
- NGA Press Release: Antineutrino Global Map Information (via NGA)
Below is a visual representation of AGM2015, illustrating the global distribution of antineutrinos:
To execute the code in this repository, you will need MATLAB version 2018a or newer. Additionally, ensure you clone and include the following dependent repositories from Ultralytics:
git clone https://github.com/ultralytics/functions-matlab
git clone https://github.com/ultralytics/nudarAfter cloning, add these repositories to your MATLAB path using the following commands:
addpath(genpath('/your_path_to/functions-matlab'));
addpath(genpath('/your_path_to/nudar'));Please ensure you have the following essential MATLAB toolboxes installed:
These toolboxes provide necessary functions for data analysis, signal processing, and geographical mapping required by the AGM2015 code. For more information on managing MATLAB environments, consult the official MathWorks documentation.
To generate the AGM2015 output using the provided code, simply execute the following command within your MATLAB environment:
fcnrunAGMThis command initiates the script that processes the data and recreates the Antineutrino Global Map based on the 2015 study. The process leverages various functions from the included repositories and toolboxes. Explore the Ultralytics documentation for more examples of scientific computing projects.
We warmly welcome contributions from the community! Your input, whether it's fixing bugs, adding new features, or improving documentation, is invaluable to us. Please see our Contributing Guide to get started. We're also keen to hear about your experiences with Ultralytics software and models; consider filling out our Survey. A huge 🙏 thank you to all our contributors for supporting our open-source initiatives! Learn more about our mission on the Ultralytics About page.
Ultralytics provides two licensing options to accommodate diverse needs:
- AGPL-3.0 License: Ideal for students and enthusiasts, this OSI-approved open-source license promotes collaboration and knowledge sharing. See the LICENSE file for details.
- Enterprise License: Designed for commercial applications, this license permits the integration of Ultralytics software and AI models into commercial products without the open-source obligations of AGPL-3.0. If your use case involves commercial deployment, please reach out through Ultralytics Licensing.
For bug reports, feature requests, and contributions, please visit GitHub Issues. For broader questions and discussions about this project or other Ultralytics work, join our vibrant community on Discord! You can also explore related topics on the Ultralytics Blog.